Service
technical service
FirePlex? miRSelect:Custom microRNA Profiling Assay
FirePlex? miRSelect is a multiplexed assay for microRNA expression analysis, allowing up to 68 small-RNA targets to be analyzed simultaneously. The assay, based on encoded hydrogel particles, can be completed in about three hours. Simply mix your mIRSelect panel with your samples in a 96-well plate, follow the streamlined protocol, and load the mixture into a flow cytometer for analysis. Software for signal normalization and expression analysis is included with every product. Scientists may also elect to have samples run at the Firefly Service Lab. Select "Run Samples at Firefly" as your Analysis Method on the form below to utilize FirePlex? microRNA analysis as a service.
《microRNA custom information table》(Click download table)
Please load websit to get more informationhttp://www.fireflybio.com
Build microRNA panel
1.1 I know my targets! (10-35 targets)
Each microRNA kit contains all reagents needed for FirePlex? miRSelect, a multiplexed assay for up to 35 microRNA targets to be quantified on standard flow cytometers. Getting started with miRSelect is easy:
1.Choose the internal controls appropriate for your experiment. If you need help, learn more about selecting controls in the FirePlex Panel Design Guide.
2.Enter up to 33 microRNA targets (miRBase v20 or custom sequences) into the form below. Two additional control targets (Blank, X-control) are automatically included with every panel for data consistency.
3.Confirm your targets and place your order.
The number of sequence reads reported in miRBase 20 will be displayed for your convenience. The number of reads does not restrict your final targets.
1.2 I know my targets! (36-68 targets)
Thank you for your interest in greater multiplexing of FirePlex miRSelect. If you are not yet sure of your microRNA targets, you can also reach Firefly through the general contact form.
Getting started with a miRSelect quote is easy:
1.Enter your contact and general panel information.
2.Choose the internal controls appropriate for your experiment. If you need help, learn more about selecting controls here.
3.Type or paste up to 68 microRNA targets (miRBase v19) into the box below. Two additional control targets (Blank, X-control) are automatically included with every panel for data consistency.
4.Confirm your targets and submit your request.
1.3 microRNA unknown

PubmiR carries your research from hypothesis to experiment in just three simple steps. Start with an exciting question and let PubmiR sift through thousands of papers in a few seconds. Use this information to build your own FirePlex? miRSelect panel and get started on microRNA research. Expand your panel as you wish and explore new frontiers. With PubmiR, you'll never miss a microRNA target again.
2,FirePlex? miRSelect performance
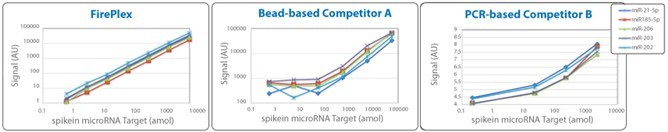
2.1FirePlex? miRSelect offers high sensitivity and broad dynamic range
FirePlex? miRSelect is a versatile microRNA assay with subattomole sensitivity (a detection limit of ~200,000 molecules) and broad dynamic range that can be used directly in cell lysate and tissue digest. The assay has been validated against qPCR, microarray, and northern blot data and is positioned to be the next generation assay for microRNA detection. Unlike many other technologies, the assay is rapid, high-throughput, and requires minimal hands-on time with few pipetting operations.
The hydrogel particles used in miRSelect enable efficient capture of your target molecules throughout the entire particle, not just on the surface like competing assays. This means you can expect consistent expression results over a broad range of assay input amount, as shown on the right.
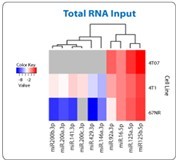
2.2 FirePlex? miRSelect has single-nucleotide specificity
miRSelect can be used for any organism and any species because of its high specificity. microRNA expression results from a total RNA extraction of C.elegans (left) show no crosstalk between microRNAs that vary by only 1-2 nucleotides.


2.3 FirePlex? miRSelect detects microRNA directly in a variety of biological samples
With miRSelect, you can expect microRNA expression results to be representative of your original sample--tumor biopsies, tissue samples, cell culture--regardless of whether you perform an RNA extraction.

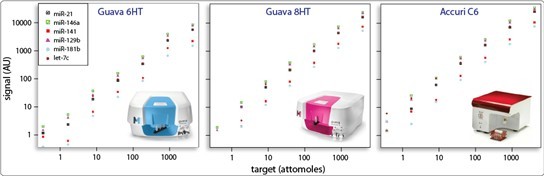
2.4 Compatibility with Standard Benchtop Cytometers
miRSelect readily expands the capabilities of almost any cytometer. We are able to support select instruments from BD, Millipore, and Miltenyi. You can read more about support for specific instruments and our expanding capabilities here.
3,Control
DESIGNING RELIABLE EXPERIMENTS To obtain reliable microRNA expression data, it is important that an experiment be designed to accurately subtract background signal, allow for appropriate normalization, and assess interwell variability. Ideally, every experiment performed with FirePlex? miRSelect includes (1) positive and blank controls in each well, (2) negative control wells, and (3) biological or technical replicates.
(1) Controls within each well
Firefly BioWorks utilizes both a positive control (“XControl”) and a blank (“Blank”) by default in each custom panel. In addition to these, it is strongly recommended that users include at least one endogenous control.
Positive control particles bear probes for a miRNA&like target, Xcontrol, that is present in Firefly Hybe Buffer at a concentration of ~1 fmol per 25 μl. This control gives confidence that the assay was successfully implemented in every well.
BLANK particles bear no probe, giving a baseline level of the background fluorescence in every assay well. This signal should be subtracted from those obtained by other targets in the same well.
Endogenous controls are small nucleolar RNAs or microRNAs expressed at consistent levels under a variety of conditions across tissues. These controls are important for signal normalization between sample types and treatments. Users are given the freedom to select endogenous controls most appropriate for their applications and may prefer to include more than one endogenous control in their panel. Firefly suggests the following endogenous controls:
Human: RNU44, RNU48, RNU6BMouse: snoRNA202, snoRNA234
Cell Lines: miR-16-5p (same sequence for human, mouse, and rat)
C. elegans: U18

(2) Negative control wells
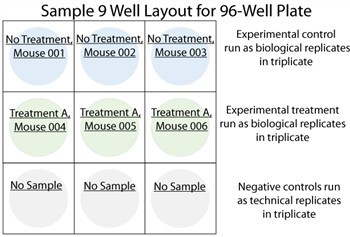
In order to obtain an accurate measurement of the background signal for each microRNA in a panel, it is necessary to run negative control wells, where carrier buffer is used in place of a biological sample. Furthermore, the use of multiple negative control wells allows users to estimate inter-well variability, giving more confidence to the results obtained. Firefly BioWorks strongly recommends the use of at least three negative controls every time an assay is performed. An example of negative control wells is provided in the sample plate layout below.
(3) Replicates
The use of replicates gives statistical meaning to results by, for example, enabling the calculation of mean and standard deviation. Replicates can be performed at the stage of sample preparation (biological) or assay (technical).
Biological replicates are those in which the same conditions are used to treat and prepare samples from different sources. These replicates are important in determining the biological variation within a population. An example of this type of replicate is serum samples derived from 3 different mice injected with the same TNF inhibitor
Technical replicates are those in which samples derived from the same source are assayed multiple times. These replicates are important in determining reproducibility of the assay. Ideally, every assay has technical replicates.